A microRNA (abbreviated miRNA) is a small non-coding RNA molecule (containing about 22 nucleotides) found in plants, animals, and some viruses, which functions in RNA silencing and post-transcriptional regulation of gene expression.
Encoded by eukaryotic nuclear DNA in plants and animals and by viral DNA in certain viruses whose genome is based on DNA, miRNAs function via base-pairing with complementary sequences within mRNA molecules. As a result, these mRNA molecules are silenced by one or more of the following processes: 1) cleavage of the mRNA strand into two pieces, 2) destabilization of the mRNA through shortening of its poly(A) tail, and 3) less efficient translation of the mRNA into proteins by ribosomes. miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short hairpins, whereas siRNAs derive from longer regions of double-stranded RNA. The human genome may encode over 1000 miRNAs, which are abundant in many mammalian cell types and appear to target about 60% of the genes of humans and other mammals.
miRNAs are well conserved in both plants and animals, and are thought to be a vital and evolutionarily ancient component of genetic regulation. While core components of the microRNA pathway are conserved between plants and animals, miRNA repertoires in the two kingdoms appear to have emerged independently with different primary modes of action. Plant miRNAs usually have near-perfect pairing with their mRNA targets, which induces gene repression through cleavage of the target transcripts. In contrast, animal miRNAs are able to recognize their target mRNAs by using as little as 6â€"8 nucleotides (the seed region) at the 5' end of the miRNA, which is not enough pairing to induce cleavage of the target mRNAs. Combinatorial regulation is a feature of miRNA regulation in animals. A given miRNA may have hundreds of different mRNA targets, and a given target might be regulated by multiple miRNAs.
The first miRNA was discovered in the early 1990s. However, miRNAs were not recognized as a distinct class of biological regulators until the early 2000s. Since then, miRNA research has revealed different sets of miRNAs expressed in different cell types and tissues and has revealed multiple roles for miRNAs in plant and animal development and in many other biological processes. Aberrant expression of miRNAs has been implicated in numerous disease states, and miRNA-based therapies are under investigation.
Estimates of the average number of unique messenger RNAs that are targets for repression by a typical microRNA vary, depending on the method used to make the estimate, but several approaches show that mammalian miRNAs can have many unique targets. For example, an analysis of the miRNAs highly conserved in vertebrate animals shows that each of these miRNAs has, on average, roughly 400 conserved targets. Likewise, experiments show that a single miRNA can reduce the stability of hundreds of unique messenger RNAs, and other experiments show that a single miRNA may repress the production of hundreds of proteins, but that this repression often is relatively mild (less than 2-fold).
History
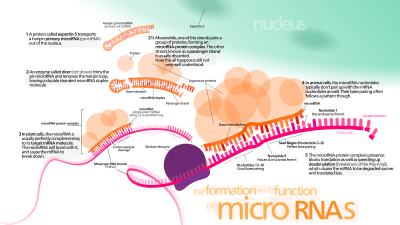
The first miRNA was discovered in 1993 by Victor Ambros, Rosalind Lee and Rhonda Feinbaum during a study of the lin-4 gene, which was known to control the timing of C. elegans larval development by repressing the lin-14 gene. When they isolated the lin-4 gene, they found that instead of producing an mRNA encoding a protein, it produced short noncoding RNAs, one of which was a ~22-nucleotide RNA that contained sequences partially complementary to multiple sequences in the 3' UTR of the lin-14 mRNA. This complementarity was proposed to inhibit the translation of the lin-14 mRNA into the LIN-14 protein. At the time, the lin-4 small RNA was thought to be a nematode idiosyncrasy. Only in 2000 was a second small RNA characterized: let-7 RNA, which represses lin-41 to promote a later developmental transition in C. elegans. The let-7 RNA was soon found to be conserved in many species, leading to the suggestion that let-7 RNA and additional "small temporal RNAs" might regulate the timing of development in diverse animals, including humans. A year later, the lin-4 and let-7 RNAs were found to be part of a very large class of small RNAs present in C. elegans, Drosophila and human cells. The many newly discovered RNAs of this class resembled the lin-4 and let-7 RNAs, except their expression patterns were usually inconsistent with a role in regulating the timing of development, which suggested that most might function in other types of regulatory pathways. At this point, researchers started using the term “microRNA†to refer to this class of small regulatory RNAs.
Nomenclature

Under a standard nomenclature system, names are assigned to experimentally confirmed miRNAs before publication of their discovery. The prefix "miR" is followed by a dash and a number, the latter often indicating order of naming. For example, miR-124 was named and likely discovered prior to miR-456. A capitalized "miR-" refers to the mature form of the miRNA, while the uncapitalized "mir-" refers to the pre-miRNA and the pri-miRNA, and "MIR" refers to the gene that encodes them. miRNAs with nearly identical sequences except for one or two nucleotides are annotated with an additional lower case letter. For example, miR-124a is closely related to miR-124b. Pre-miRNAs, pri-miRNAs and genes that lead to 100% identical mature miRNAs but that are located at different places in the genome are indicated with an additional dash-number suffix. For example, the pre-miRNAs hsa-mir-194-1 and hsa-mir-194-2 lead to an identical mature miRNA (hsa-miR-194) but are from genes located in different regions of the genome. Species of origin is designated with a three-letter prefix, e.g., hsa-miR-124 is a human (Homo sapiens) miRNA and oar-miR-124 is a sheep (Ovis aries) miRNA. Other common prefixes include 'v' for viral (miRNA encoded by a viral genome) and 'd' for Drosophila miRNA (a fruit fly commonly studied in genetic research). When two mature microRNAs originate from opposite arms of the same pre-miRNA and are found in roughly similar amounts, they are denoted with a -3p or -5p suffix. (In the past, this distinction was also made with 's' (sense) and 'as' (antisense)). However, the mature microRNA found from one arm of the hairpin is usually much more abundant than that found from the other arm, in which case, an asterisk following the name indicates the mature species found at low levels from the opposite arm of a hairpin. For example, miR-124 and miR-124* share a pre-miRNA hairpin, but much more miR-124 is found in the cell.
Biogenesis

MicroRNAs are produced from either their own genes or from introns. A video of this process can be found here.
The majority of the characterized miRNA genes are intergenic or oriented antisense to neighboring genes and are therefore suspected to be transcribed as independent units. However, in some cases a microRNA gene is transcribed together with its host gene; this provides a means for coupled regulation of miRNA and protein-coding gene. As much as 40% of miRNA genes may lie in the introns of protein and non-protein coding genes or even in exons of long nonprotein-coding transcripts. These are usually, though not exclusively, found in a sense orientation, and thus usually are regulated together with their host genes. Other miRNA genes showing a common promoter include the 42-48% of all miRNAs originating from polycistronic units containing multiple discrete loops from which mature miRNAs are processed, although this does not necessarily mean the mature miRNAs of a family will be homologous in structure and function. The promoters mentioned have been shown to have some similarities in their motifs to promoters of other genes transcribed by RNA polymerase II such as protein coding genes. The DNA template is not the final word on mature miRNA production: 6% of human miRNAs show RNA editing (IsomiRs), the site-specific modification of RNA sequences to yield products different from those encoded by their DNA. This increases the diversity and scope of miRNA action beyond that implicated from the genome alone.
Transcription
miRNA genes are usually transcribed by RNA polymerase II (Pol II). The polymerase often binds to a promoter found near the DNA sequence encoding what will become the hairpin loop of the pre-miRNA. The resulting transcript is capped with a specially modified nucleotide at the 5’ end, polyadenylated with multiple adenosines (a poly(A) tail), and spliced. Animal miRNAs are initially transcribed as part of one arm of an ∼80 nucleotide RNA stem-loop that in turn forms part of a several hundred nucleotides long miRNA precursor termed a primary miRNA (pri-miRNA)s. When a stem-loop precursor is found in the 3' UTR, a transcript may serve as a pri-miRNA and a mRNA. RNA polymerase III (Pol III) transcribes some miRNAs, especially those with upstream Alu sequences, transfer RNAs (tRNAs), and mammalian wide interspersed repeat (MWIR) promoter units.
Nuclear processing
A single pri-miRNA may contain from one to six miRNA precursors. These hairpin loop structures are composed of about 70 nucleotides each. Each hairpin is flanked by sequences necessary for efficient processing. The double-stranded RNA structure of the hairpins in a pri-miRNA is recognized by a nuclear protein known as DiGeorge Syndrome Critical Region 8 (DGCR8 or "Pasha" in invertebrates), named for its association with DiGeorge Syndrome. DGCR8 associates with the enzyme Drosha, a protein that cuts RNA, to form the "Microprocessor" complex. In this complex, DGCR8 orients the catalytic RNase III domain of Drosha to liberate hairpins from pri-miRNAs by cleaving RNA about eleven nucleotides from the hairpin base (two helical RNA turns into the stem). The product resulting has a two-nucleotide overhang at its 3’ end; it has 3' hydroxyl and 5' phosphate groups. It is often termed as a pre-miRNA (precursor-miRNA).
Pre-miRNAs that are spliced directly out of introns, bypassing the Microprocessor complex, are known as "Mirtrons." Originally thought to exist only in Drosophila and C. elegans, mirtrons have now been found in mammals.
Perhaps as many as 16% of pre-miRNAs may be altered through nuclear RNA editing. Most commonly, enzymes known as adenosine deaminases acting on RNA (ADARs) catalyze adenosine to inosine (A to I) transitions. RNA editing can halt nuclear processing (for example, of pri-miR-142, leading to degradation by the ribonuclease Tudor-SN) and alter downstream processes including cytoplasmic miRNA processing and target specificity (e.g., by changing the seed region of miR-376 in the central nervous system).
Nuclear export
Pre-miRNA hairpins are exported out of the nucleus in a process involving the nucleocytoplasmic shuttler Exportin-5. This protein, a member of the karyopherin family, recognizes a two-nucleotide overhang left by the RNase III enzyme Drosha at the 3' end of the pre-miRNA hairpin. Exportin-5-mediated transport to the cytoplasm is energy-dependent, using GTP bound to the Ran protein.
Cytoplasmic processing
In the cytoplasm, the pre-miRNA hairpin is cleaved by the RNase III enzyme Dicer. This endoribonuclease interacts with the 3' end of the hairpin and cuts away the loop joining the 3' and 5' arms, yielding an imperfect miRNA:miRNA* duplex about 22 nucleotides in length. Overall hairpin length and loop size influence the efficiency of Dicer processing, and the imperfect nature of the miRNA:miRNA* pairing also affects cleavage. Although either strand of the duplex may potentially act as a functional miRNA, only one strand is usually incorporated into the RNA-induced silencing complex (RISC) where the miRNA and its mRNA target interact.
Biogenesis in plants
miRNA biogenesis in plants differs from animal biogenesis mainly in the steps of nuclear processing and export. Instead of being cleaved by two different enzymes, once inside and once outside the nucleus, both cleavages of the plant miRNA is performed by a Dicer homolog, called Dicer-like1 (DL1). DL1 is only expressed in the nucleus of plant cells, which indicates that both reactions take place inside the nucleus. Before plant miRNA:miRNA* duplexes are transported out of the nucleus, its 3' overhangs are methylated by a RNA methyltransferaseprotein called Hua-Enhancer1 (HEN1). The duplex is then transported out of the nucleus to the cytoplasm by a protein called Hasty (HST), an Exportin 5 homolog, where they disassemble and the mature miRNA is incorporated into the RISC.
The RNA-induced silencing complex
The mature miRNA is part of an active RNA-induced silencing complex (RISC) containing Dicer and many associated proteins. RISC is also known as a microRNA ribonucleoprotein complex (miRNP); RISC with incorporated miRNA is sometimes referred to as "miRISC."
Dicer processing of the pre-miRNA is thought to be coupled with unwinding of the duplex. Generally, only one strand is incorporated into the miRISC, selected on the basis of its thermodynamic instability and weaker base-pairing relative to the other strand. The position of the stem-loop may also influence strand choice. The other strand, called the passenger strand due to its lower levels in the steady state, is denoted with an asterisk (*) and is normally degraded. In some cases, both strands of the duplex are viable and become functional miRNA that target different mRNA populations.
Members of the Argonaute (Ago) protein family are central to RISC function. Argonautes are needed for miRNA-induced silencing and contain two conserved RNA binding domains: a PAZ domain that can bind the single stranded 3’ end of the mature miRNA and a PIWI domain that structurally resembles ribonuclease-H and functions to interact with the 5’ end of the guide strand. They bind the mature miRNA and orient it for interaction with a target mRNA. Some argonautes, for example human Ago2, cleave target transcripts directly; argonautes may also recruit additional proteins to achieve translational repression. The human genome encodes eight argonaute proteins divided by sequence similarities into two families: AGO (with four members present in all mammalian cells and called E1F2C/hAgo in humans), and PIWI (found in the germ line and hematopoietic stem cells).
Additional RISC components include TRBP [human immunodeficiency virus (HIV) transactivating response RNA (TAR) binding protein], PACT (protein activator of the interferon induced protein kinase (PACT), the SMN complex, fragile X mental retardation protein (FMRP), Tudor staphylococcal nuclease-domain-containing protein (Tudor-SN), the putative DNA helicase MOV10, and the RNA recognition motif containing protein TNRC6B.
Mode of silencing
Gene silencing may occur either via mRNA degradation or preventing mRNA from being translated. For example, miR16 contains a sequence complementary to the AU-rich element found in the 3'UTR of many unstable mRNAs, such as TNF alpha or GM-CSF (ref Jing, Q., et al. Cell 2005 pmid = 15766526). It has been demonstrated that if there is complete complementation between the miRNA and target mRNA sequence, Ago2 can cleave the mRNA and lead to direct mRNA degradation. Yet, if there isn't complete complementation the silencing is achieved by preventing translation.
miRNA turnover
Turnover of mature miRNA is needed for rapid changes in miRNA expression profiles. During miRNA maturation in the cytoplasm, uptake by the Argonaute protein is thought to stabilize the guide strand, while the opposite (* or "passenger") strand is preferentially destroyed. In what has been called a "Use it or lose it" strategy, Argonaute may preferentially retain miRNAs with many targets over miRNAs with few or no targets, leading to degradation of the non-targeting molecules.
Decay of mature miRNAs in Caenorhabditis elegans is mediated by the 5´-to-3´ exoribonuclease XRN2, also known as Rat1p. In plants, SDN (small RNA degrading nuclease) family members degrade miRNAs in the opposite (3'-to-5') direction. Similar enzymes are encoded in animal genomes, but their roles have not yet been described.
Several miRNA modifications affect miRNA stability. As indicated by work in the model organism Arabidopsis thaliana (thale cress), mature plant miRNAs appear to be stabilized by the addition of methyl moieties at the 3' end. The 2'-O-conjugated methyl groups block the addition of uracil (U) residues by uridyltransferase enzymes, a modification that may be associated with miRNA degradation. However, uridylation may also protect some miRNAs; the consequences of this modification are incompletely understood. Uridylation of some animal miRNAs has also been reported. Both plant and animal miRNAs may be altered by addition of adenine (A) residues to the 3' end of the miRNA. An extra A added to the end of mammalian miR-122, a liver-enriched miRNA important in Hepatitis C, stabilizes the molecule, and plant miRNAs ending with an adenine residue have slower decay rates.
Cellular functions

The function of miRNAs appears to be in gene regulation. For that purpose, a miRNA is complementary to a part of one or more messenger RNAs (mRNAs). Animal miRNAs are usually complementary to a site in the 3' UTR whereas plant miRNAs are usually complementary to coding regions of mRNAs. Perfect or near perfect base pairing with the target RNA promotes cleavage of the RNA. This is the primary mode of plant miRNAs. In animals miRNAs more often have only partly the right sequence of nucleotides to bond with the target mRNA. The match-ups are imperfect. For partially complementary microRNAs to recognise their targets, nucleotides 2â€"7 of the miRNA (its 'seed region') still have to be perfectly complementary. Animal miRNAs inhibit protein translation of the target mRNA (this exists in plants as well but is less common). MicroRNAs that are partially complementary to a target can also speed up deadenylation, causing mRNAs to be degraded sooner. While degradation of miRNA-targeted mRNA is well documented, whether or not translational repression is accomplished through mRNA degradation, translational inhibition, or a combination of the two is hotly debated. Recent work on miR-430 in zebrafish, as well as on bantam-miRNA and miR-9 in Drosophila cultured cells, shows that translational repression is caused by the disruption of translation initiation, independent of mRNA deadenylation.
miRNAs occasionally also cause histone modification and DNA methylation of promoter sites, which affects the expression of target genes.
Nine mechanisms of miRNA action are described and assembled in a unified mathematical model:
- Cap-40S initiation inhibition;
- 60S Ribosomal unit joining inhibition;
- Elongation inhibition;
- Ribosome drop-off (premature termination);
- Co-translational nascent protein degradation;
- Sequestration in P-bodies;
- mRNA Decay (destabilisation);
- mRNA Cleavage;
- Transcriptional inhibition through microRNA-mediated chromatin reorganization following by gene silencing.
It is often impossible to discern these mechanisms using the experimental data about stationary reaction rates. Nevertheless, they are differentiated in dynamics and have different kinetic signatures.
Unlike plant microRNAs, the animal microRNAs target a diverse set of genes. However, genes involved in functions common to all cells, such as gene expression, have relatively fewer microRNA target sites and seem to be under selection to avoid targeting by microRNAs.
dsRNA can also activate gene expression, a mechanism that has been termed "small RNA-induced gene activation" or RNAa. dsRNAs targeting gene promoters can induce potent transcriptional activation of associated genes. This was demonstrated in human cells using synthetic dsRNAs termed small activating RNAs (saRNAs), but has also been demonstrated for endogenous microRNA.
Interactions between microRNAs and complementary sequences on genes and even pseudogenes that share sequence homology are thought to be a back channel of communication regulating expression levels between paralogous genes. Given the name "competing endogenous RNAs" (ceRNAs), these microRNAs bind to "microRNA response elements" on genes and pseudogenes and may provide another explanation for the persistence of non-coding DNA.
Evolution
MicroRNAs are significant phylogenetic markers because of their astonishingly low rate of evolution. MicroRNAs origin as a regulatory mechanism developed from previous RNAi machinery which was initially used as a defense against exogenous genetic material such as viruses. Their origin may have permitted the development of morphological innovation, and by making gene expression more specific and 'fine-tunable', permitted the genesis of complex organs and perhaps, ultimately, complex life. Indeed, rapid bursts of morphological innovation are generally associated with a high rate of microRNA accumulation.
New microRNAs are created in multiple different ways. Novel microRNAs can originate from the random formation of hairpins in "non-coding" sections of DNA (i.e. introns or intergene regions), but also by the duplication and modification of existing microRNAs. MicroRNAs can also form from inverted duplications of protein-coding sequences, which allows for the creation of a foldback hairpin structure. The rate of evolution (i.e. nucleotide substitution) in recently originated microRNAs is comparable to that elsewhere in the non-coding DNA, implying evolution by neutral drift; however, older microRNAs have a much lower rate of change (often less than one substitution per hundred million years), suggesting that once a microRNA gains a function it undergoes extreme purifying selection. Additionally, different regions within an miRNA gene seem to be under different evolutionary pressures, where regions that are vital for processing and function have much higher levels of conservation. At this point, a microRNA is rarely lost from an animal's genome, although microRNAs that are more recently derived (and thus presumably non-functional) are frequently lost. In Arabidopsis thaliana, the net flux of miRNA genes has been predicted to be between 1.2 and 3.3 genes per million years. This makes them a valuable phylogenetic marker, and they are being looked upon as a possible solution to such outstanding phylogenetic problems as the relationships of arthropods.
MicroRNAs feature in the genomes of most eukaryotic organisms, from the brown algae to the animals. However, the difference in how these microRNAs function and the way they are processed suggests that microRNAs arose independently in plants and animals. Focusing on the animals, the genome of Mnemiopsis leidyi appears to lack recognizable microRNAs, as well as the nuclear proteins Drosha and Pasha, which are critical to canonical microRNA biogenesis. It is the only animal thus far reported to be missing Drosha. MicroRNAs play a vital role in the regulation of gene expression in all non-ctenophore animals investigated thus far except for Trichoplax adhaerens, the only known member of the phylum Placozoa.
Across all species, in excess of 5000 had been identified by March 2010. Whilst short RNA sequences (50 â€" hundreds of base pairs) of a broadly comparable function occur in bacteria, bacteria lack true microRNAs.
Experimental detection and manipulation of miRNA
While researchers have focused on the study of miRNA expression in physiological and pathological processes, various technical variables related to microRNA isolation have emerged. The stability of the stored miRNA samples has often been questioned. MicroRNAs are degraded much more easily than mRNAs, partly due to their length, but also because of the ubiquitously present RNases. This makes it necessary to cool samples on ice and use RNase-free equipment whenever working with microRNAs.
MicroRNA expression can be quantified in a two-step polymerase chain reaction process of modified RT-PCR followed by quantitative PCR. Variations of this method achieve absolute or relative quantification. miRNAs can also be hybridized to microarrays, slides or chips with probes to hundreds or thousands of miRNA targets, so that relative levels of miRNAs can be determined in different samples. MicroRNAs can be both discovered and profiled by high-throughput sequencing methods (MicroRNA Sequencing). The activity of an miRNA can be experimentally inhibited using a locked nucleic acid (LNA) oligo, a Morpholino oligo or a 2'-O-methyl RNA oligo. Additionally, a specific miRNA can be silenced by a complementary antagomir. MicroRNA maturation can be inhibited at several points by steric-blocking oligos. The miRNA target site of an mRNA transcript can also be blocked by a steric-blocking oligo. For the “in situ†detection of miRNA, LNA or Morpholino probes can be used. The locked conformation of LNA results in enhanced hybridization properties and increases sensitivity and selectivity, making it ideal for detection of short miRNA.
High-throughput quantification of miRNAs is often difficult and prone to errors, for the larger variance (compared to mRNAs) that comes with the methodological problems. mRNA-expression is therefore often analyzed as well to check for miRNA-effects in their levels (e. g. in ). To pair mRNA- and miRNA-data, databases can be used which predict miRNA-targets based on their base sequence. While this is usually done after miRNAs of interest have been detected (e. g. because of high expression levels), ideas for analysis tools that integrate mRNA- and miRNA-expression information have been proposed.
Disease
Just as miRNA is involved in the normal functioning of eukaryotic cells, so has dysregulation of miRNA been associated with disease. A manually curated, publicly available database, miR2Disease, documents known relationships between miRNA dysregulation and human disease.
Inherited diseases
A mutation in the seed region of miR-96, causes hereditary progressive hearing loss.
A mutation in the seed region of miR-184, causes hereditary keratoconus with anterior polar cataract.
Deletion of the miR-17~92 cluster, causes skeletal and growth defects.
Cancer
The first human disease known to be associated with miRNA deregulation was chronic lymphocytic leukemia. Many miRNAs have subsequently been found to have links with various types of cancer and accordingly are sometimes referred to as "oncomirs". MicroRNA-21 is involved in several cancer-types such as glioblastoma and astrocytoma was one of the first microRNAs to be identified as an oncomir.
A study of mice altered to produce excess c-Myc â€" a protein with mutated forms implicated in several cancers â€" shows that miRNA has an effect on the development of cancer. Mice that were engineered to produce a surplus of types of miRNA found in lymphoma cells developed the disease within 50 days and died two weeks later. In contrast, mice without the surplus miRNA lived over 100 days. Leukemia can be caused by the insertion of a viral genome next to the 17-92 array of microRNAs leading to increased expression of this microRNA.
Another study found that two types of miRNA inhibit the E2F1 protein, which regulates cell proliferation. miRNA appears to bind to messenger RNA before it can be translated to proteins that switch genes on and off.
By measuring activity among 217 genes encoding miRNA, patterns of gene activity that can distinguish types of cancers can be discerned. miRNA signatures may enable classification of cancer. This will allow doctors to determine the original tissue type which spawned a cancer and to be able to target a treatment course based on the original tissue type. miRNA profiling has already been able to determine whether patients with chronic lymphocytic leukemia had slow growing or aggressive forms of the cancer.
Transgenic mice that over-express or lack specific miRNAs have provided insight into the role of small RNAs in various malignancies. Much work has also been done on the role of microRNAs in establishing and maintaining cancer stem cells that are especially resistant to chemotherapy and often responsible for relapse.
A novel miRNA-profiling based screening assay for the detection of early-stage colorectal cancer has been developed and is currently in clinical trials. Early results showed that blood plasma samples collected from patients with early, resectable (Stage II) colorectal cancer could be distinguished from those of sex-and age-matched healthy volunteers. Sufficient selectivity and specificity could be achieved using small (less than 1 mL) samples of blood. The test has potential to be a cost-effective, non-invasive way to identify at-risk patients who should undergo colonoscopy.
Another role for miRNA in cancers is to use their expression level as a prognostic. For example, one study on NSCLC samples found that low miR-324a levels could serve as a prognostic indicator of poor survival. Either high miR-185 or low miR-133b levels may correlate with metastasis and poor survival in colorectal cancer. Hepatocellular carcinoma cell proliferation may arise from miR-21 interaction with MAP2K3, a tumor repressor gene. Optimal treatment for cancer involves accurately identifying patients for risk-stratified therapy. Those with a rapid response to initial treatment may benefit from truncated treatment regimens, thus the need for more accurate measures of disease response. Cell-free miRNA are highly stable in blood, are overexpressed in cancer, and are quantifiable within the diagnostic laboratory. In classical Hodgkin lymphoma, plasma miR-21, miR-494, and miR-1973 are promising disease response biomarkers. Circulating miRNAs have the potential to greatly assist clinical decision making and aid interpretation of positron emission tomography combined with computerized tomography. A further advantage is they can also be performed at each consultation to assess disease response and detection of early relapse.
Recent studies have miR-205 targeted for inhibiting the metastatic nature of breast cancer. Five members of the microRNA-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) are down regulated in tumour progression of breast cancer.
DNA repair and cancer
DNA damage is considered to be the primary underlying cause of cancer. If DNA repair is deficient, damage tends to accumulate in DNA. Such DNA damage can cause mutational errors during DNA replication due to error-prone translesion synthesis. Accumulated DNA damage can also cause epigenetic alterations due to errors during DNA repair. Such mutations and epigenetic alterations can give rise to cancer (see malignant neoplasms).
Germ line mutations in DNA repair genes cause only 2â€"5% of colon cancer cases. However, altered expression of microRNAs, causing DNA repair deficiencies, are frequently associated with cancers and may be an important causal factor for these cancers.
Among 68 sporadic colon cancers with reduced expression of the DNA mismatch repair protein MLH1, most were found to be deficient due to epigenetic methylation of the CpG island of the MLH1 gene. However, up to 15% of the MLH1-deficiencies in sporadic colon cancers appeared to be due to over-expression of the microRNA miR-155, which represses MLH1 expression.
In 29â€"66% of glioblastomas, DNA repair is deficient due to epigenetic methylation of the MGMT gene, which reduces protein expression of MGMT. However, for 28% of glioblastomas, the MGMT protein is deficient but the MGMT promoter is not methylated. In the glioblastomas without methylated MGMT promoters, the level of microRNA miR-181d is inversely correlated with protein expression of MGMT and the direct target of miR-181d is the MGMT mRNA 3’UTR (the three prime untranslated region of MGMT mRNA). Thus, in 28% of glioblastomas, increased expression of miR-181d and reduced expression of DNA repair enzyme MGMT may be a causal factor.
HMGA proteins (HMGA1a, HMGA1b and HMGA2) are implicated in cancer, and expression of these proteins is regulated by microRNAs. HMGA expression is almost undetectable in differentiated adult tissues but is elevated in many cancers. HGMA proteins are polypeptides of ~100 amino acid residues characterized by a modular sequence organization. These proteins have three highly positively-charged regions, termed AT hooks, that bind the minor groove of AT-rich DNA stretches in specific regions of DNA. Human neoplasias, including thyroid, prostatic, cervical, colorectal, pancreatic and ovarian carcinoma, show a strong increase of HMGA1a and HMGA1b proteins. Transgenic mice with HMGA1 targeted to lymphoid cells develop aggressive lymphoma, showing that high HMGA1 expression is not only associated with cancers, but that the HMGA1 gene can act as an oncogene to cause cancer. Baldassarre et al., showed that HMGA1 protein binds to the promoter region of DNA repair gene BRCA1 and inhibits BRCA1 promoter activity. They also showed that while only 11% of breast tumors had hypermethylation of the BRCA1 gene, 82% of aggressive breast cancers have low BRCA1 protein expression, and most of these reductions were due to chromatin remodeling by high levels of HMGA1 protein.
HMGA2 protein specifically targets the promoter of ERCC1, thus reducing expression of this DNA repair gene. ERCC1 protein expression was deficient in 100% of 47 evaluated colon cancers (though the extent to which HGMA2 was involved is not known). Palmieri et al. showed that, in normal tissues, HGMA1 and HMGA2 genes are targeted (and thus strongly reduced in expression) by miR-15, miR-16, miR-26a, miR-196a2 and Let-7a. However, each of these HMGA-targeting miRNAs are drastically reduced in almost all human pituitary adenomas studied, when compared with the normal pituitary gland. Consistent with the down-regulation of these HMGA-targeting miRNAs, an increase in the HMGA1 and HMGA2-specific mRNAs was observed. Three of these microRNAs (miR-16, miR-196a and Let-7a) have methylated promoters and therefore low expression in colon cancer. For two of these, miR-15 and miR-16, the coding regions are epigenetically silenced in cancer due to histone deacetylase activity. When these microRNAs are expressed at a low level, then HMGA1 and HMGA2 proteins are expressed at a high level. HMGA1 and HMGA2 target (reduce expression of) BRCA1 and ERCC1 DNA repair genes. Thus DNA repair can be reduced, likely contributing to cancer progression.
In contrast to the previous example, where under-expression of miRNAs indirectly caused reduced expression of DNA repair genes, in some cases over-expression of certain miRNAs may directly reduce expression of specific DNA repair proteins. Wan et al. referred to 6 DNA repair genes that are directly targeted by the miRNAs indicated: ATM (miR-421), RAD52 (miR-210, miR-373), RAD23B (miR-373), MSH2 (miR-21), BRCA1 (miR-182) and P53 (miR-504, miR-125b). More recently, Tessitore et al. listed multiple DNA repair genes directly targeted by these additional miRNAs: ATM (miR-100, miR18a, miR-101), DNA-PK (miR-101), ATR (mir-185), Wip1 (miR-16), MLH1, MSH2, MSH6 (miR-155), ERCC3, ERCC4 (miR-192) and UNG2 (miR-16, miR-34c). Among these miRNAs, miR-16, miR-18a, miR-21, miR-34c, miR-101, miR-125b, miR-155, miR-182, miR-185, miR-192 and miR-373 were identified by Schnekenburger and Diederich as over-expressed in colon cancer through epigenetic hypomethylation. Over expression of any one of these miRNAs can cause reduced expression of its target DNA repair gene.
Heart disease
The global role of miRNA function in the heart has been addressed by conditionally inhibiting miRNA maturation in the murine heart, and has revealed that miRNAs play an essential role during its development. miRNA expression profiling studies demonstrate that expression levels of specific miRNAs change in diseased human hearts, pointing to their involvement in cardiomyopathies. Furthermore, studies on specific miRNAs in animal models have identified distinct roles for miRNAs both during heart development and under pathological conditions, including the regulation of key factors important for cardiogenesis, the hypertrophic growth response, and cardiac conductance.
miRNA-712
Murine microRNA-712 is a potential biomarker (i.e. predictor) for atherosclerosis, a cardiovascular disease of the arterial wall associated with lipid retention and inflammation. Non-laminar blood flow also correlates with development of atherosclerosis as mechanosenors of epithelial cells respond to the sheer force of disturbed flow (d-flow). A number of pro-atherogenic genes including matrix metalloproteinases (MMPs) are upregulated by d-flow , mediating pro-inflammatory and pro-angiogenic signals. These findings were observed in ligated carotid arteries of mice to mimic the effects of d-flow. Within 24 hours, pre-existing immature miR-712 formed mature miR-712 suggesting that miR-712 is flow-sensitive. Coinciding with these results, miR-712 is also upregulated in endothelial cells exposed to naturally occurring d-flow in the greater curvature of the aortic arch.
Gene Origin
Pre-mRNA sequence of miR-712 is generated from the murine ribosomal RN45s gene at the internal transcribed spacer region 2 (ITS2). XRN1 is an exonuclease that degrades the ITS2 region during processing of RN45s . Reduction of XRN1 under d-flow conditions therefore leads to the accumulation of miR-712.
Mechanism
MiR-712 targets tissue inhibitor of metalloproteinases 3 (TIMP3). TIMPs normally regulate activity of matrix metalloproteinases (MMPs) which degrade the extracellular matrix (ECM). Arterial ECM is mainly composed of collagen and elastin fibers, providing the structural support and recoil properties of arteries. These fibers play a critical role in regulation of vascular inflammation and permeability, which are important in the development of atherosclerosis. Expressed by endothelial cells, TIMP3 is the only ECM bound TIMP. A decrease in TIMP3 expression results in an increase of ECM degradation in the presence of d-flow. Consistent with these findings, inhibition of pre-miR712 increases expression of TIMP3 in cells, even when exposed to turbulent flow.
TIMP3 also decreases the expression of TNFα (a pro-inflammatory regulator) during turbulent flow. Activity of TNFα in turbulent flow was measured by the expression of TNFα converting enzyme (TACE) in blood. TNFα decreased if miR-712 was inhibited or TIMP3 overexpressed, suggesting that miR-712 and TIMP3 regulate TACE activity in turbulent flow conditions.
Anti-miR-712 effectively suppresses d-flow induced miR-712 expression and increases TIMP3 expression. Anti-miR-712 also inhibits vascular hyperpermeability, thereby significantly reducing atherosclerosis lesion development and immune cell infiltration.
Human Homolog microRNA-205
The human homolog of miR-712 was found on the RN45s homolog gene, which maintains similar miRNAs to mice. MiR-205 of humans share similar sequences with miR-712 of mice and is conserved across most vertebrates. MiR-205 and miR-712 also share more than 50% of the cell signaling targets, including TIMP3.
When tested, d-flow decreased the expression of XRN1 in humans as it did in mice endothelial cells, indicating that there may be a common role of XRN1 in human. While the human homolog has not been thoroughly studied, the discovery and function of miRNA-712 can give weight for future research on its potential as a biomarker in mice models of atherosclerosis.
Nervous system
miRNAs appear to regulate the nervous system. Neural miRNAs are involved at various stages of synaptic development, including dendritogenesis (involving miR-132, miR-134 and miR-124), synapse formation and synapse maturation (where miR-134 and miR-138 are thought to be involved). Some studies find altered miRNA expression in schizophrenia.
Obesity
miRNAs play crucial roles in the regulation of stem cell progenitors differentiating into adipocytes. Studies to determine what role pluripotent stem cells play in adipogenesis, were examined in the immortalized human bone marrow-derived stromal cell line hMSC-Tert20. Decreased expression of miR-155,miR-221,and miR-222, have been found during the adipogenic programming of both immortalized and primary hMSCs, suggesting that they act as negative regulators of differentiation. Conversely, ectopic expression of the miRNAs 155,221, and 222 significantly inhibited adipogenesis and repressed induction of the master regulators PPARγ and CCAAT/enhancer-binding protein alpha (CEBPA). This paves the way for possible obesity treatments on the genetic level.
Another class of miRNAs that regulate insulin resistance, obesity, and diabetes, is the let-7 family. Let-7 is known to accumulate in human tissues during the course of aging. When let-7 was ectopically overexpressed to mimic accelerated aging, mice became insulin-resistant, and thus more prone to high fat diet-induced obesity and diabetes. In contrast when let-7 was inhibited by injections of let-7-specific antagomirs, mice become more insulin-sensitive, and remarkably resistant to high fat diet-induced obesity and diabetes. Not only could let-7 inhibition prevent obesity and diabetes, it could also reverse and cure diabetes. These experimental findings suggest that let-7 inhibition could represent a new therapy for obesity and type 2 diabetes.
Non-coding RNAs

When the human genome project mapped its first chromosome in 1999, it was predicted the genome would contain over 100,000 protein coding genes. However, only around 20,000 were eventually identified (International Human Genome Sequencing Consortium, 2004). Since then, the advent of bioinformatics approaches combined with genome tiling studies examining the transcriptome, systematic sequencing of full length cDNA libraries, and experimental validation (including the creation of miRNA derived antisense oligonucleotides called antagomirs) have revealed that many transcripts are non protein-coding RNA, including several snoRNAs and miRNAs.
Viruses
The expression of transcription activators by human herpesvirus-6 DNA, is believed to be regulated by viral miRNA.
Target prediction
miRNAs can bind to target messenger RNA (mRNA) transcripts of protein-coding genes and negatively control their translation or cause mRNA degradation. It is of key importance to identify the miRNA targets accurately. A detailed review for the advances in the miRNA target identification methods and available resources has been published by Zheng et al.
